Genes vs Proteins: A perspective from brain tumor research
Aniruddha Mukherjee
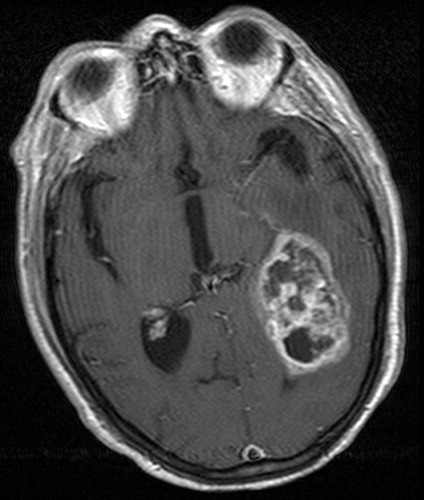 Brain scan showing enhancing GBM tumor mass. Courtesy:
RadioGraphics, Radiological Society of North America.
Brain scan showing enhancing GBM tumor mass. Courtesy:
RadioGraphics, Radiological Society of North America.
The plethora of molecules inside a cell is highly regulated
in terms of their abundance and functions. The regulation
happens in layers which are, again, most fundamentally
attributed to the expression of genes and proteins. The
present article enlightens us on how a large-scale the study
of proteins and genes in one of the deadliest tumors reveals
layer-specific association of molecules with the time of
survival of patients.
Glioblastoma multiforme (GBM) is an aggressive
form of brain tumor with incidences ranging from 1 to 5 out
of 100000 people. The average survival of patients with such
tumors is less than two years. Many new treatment therapies
and drugs have recently shown promise; however, the caveat
remains in understanding the heterogeneity of these tumors
at the molecular level. Tumor heterogeneity refers to the
differences in the same type of tumor across different
patients that can arise due to multiple causes like the
types of molecules in the tumor, time of preserving clinical
samples, and the lifestyle of patients, etc. Eventually,
tumor heterogeneity becomes a reason for varying responses
to treatment.
A group of scientists from Tel Aviv University, Israel, have
recently forged their way into GBM research showing clear
demarcation between the fundamental biological molecules in
terms of their ability to correlate with patient-specific
characteristic features and find how proteins can
distinctively inform better about certain features. One of
the modern methods used by them in this work is “Proteomics,
'' a broad term in research where the entire protein
complement of a cell, tissue or organ is analyzed to extract
information about the concerned condition. Simultaneously,
they also studied the mRNA, a molecule consisting of the
gene and serves as the foundation for protein synthesis
inside the cell. The term coined to encompass all the mRNA
molecules in a certain cell or tissue of an organism is
‘transcriptome’. Their research tells us how studying the
proteome( i.e., the whole protein expressed in an organism
at a certain time) inside the tumor tissues can turn out to
be more suggestive of GBM patient prognosis compared to the
information obtained exclusively from the genes. The
intricacies in the process of protein synthesis gives rise
to a higher number of total proteins which are in a constant
state of interaction , hence making the proteome enormously
complex and dynamic. This is likely to be a reason why
proteomics by itself has become an indispensable research
tool in medicine.
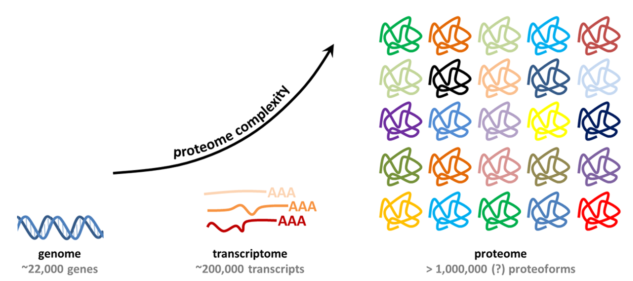 Image credits: Proteomics Center, Erasmus University
Medical Center.
Image credits: Proteomics Center, Erasmus University
Medical Center.
Tamar Geiger and his team of scientists, selected a cohort
of 87 patients, out of which 22 were processed to study the
proteome, 33 samples were used for sequencing the mRNA,
while 32 were subjected to analysis at both levels of
proteome and transcriptome. Here, the researchers have taken
a routine approach of extracting the whole RNA or protein
from the tumor tissues and subjecting those to analytical
platforms to generate huge amounts of data in terms of the
types, abundance and interaction of each molecule.
Subsequent analysis of this information helped draw relevant
conclusions about a smaller set of molecules being
associated with the specific biological processes which are
likely to go wrong in a tumor cell. For instance,neuron
generation is one such process enriched by both proteins and
genes, suggesting how there is an expansion of the formation
of nerve cells or neurons contributing to the prevalence of
GBM in the brain. Similarly, there are clinical parameters
which consist of the data of tumor location inside the
brain, patient survival after onset, tumor recurrence after
treatment of patients etc. After analyzing the tumor
samples, the researchers arrived at a set of significant
genes and proteins and noticed that the proteins could
independently show a strong association with the clinical
data of patient survival on a statistical basis, whereas
genes did not correlate at all. This particular analysis saw
an extended version as researchers integrated both proteome
and transcriptome-based data and then tried to correlate
with survival bifurcated into a binary of short (less than 6
months) and long (more than 2 years) time of GBM patients.
This eventually produced results of genes and proteins
showing similar correlation only when compared with data
from shorter survival! The researchers have clearly
communicated that because their study imbibded high
heterogeneity with mutations in the GBM genes, there is a
future scope for extrapolating similar research with even
larger cohorts of patients. Nevertheless, this study puts
out multiple aberrant biological processes in GBM and shows
how research targeted to specific layers of molecules can
tackle the existing challenge of tumor heterogeneity to an
extent that reveal the most novel molecular associations.
References
-
Yanovich-Arad, Gali, Paula Ofek, Eilam Yeini, Mariya
Mardamshina, Artem Danilevsky, Noam Shomron, Rachel
Grossman, Ronit Satchi-Fainaro, and Tamar Geiger.
“Proteogenomics of Glioblastoma Associates Molecular
Patterns with Survival.” Cell Reports 34, no. 9 (March
2, 2021): 108787.
Aniruddha Mukherjee completed his BS-MS with a major in
Biological Sciences from IISER Kolkata. He will be joining
University of Alabama at Birmingham, U.S.A for his graduate
studies in Biomedical Sciences in the coming fall. Apart from
being a movie buff he enjoys cooking and playing chess.
 Brain scan showing enhancing GBM tumor mass. Courtesy:
RadioGraphics, Radiological Society of North America.
Brain scan showing enhancing GBM tumor mass. Courtesy:
RadioGraphics, Radiological Society of North America.
